Note: This blog originally appeared on the Nature Portfolio Microbiology Community.
Typhoid fever is a serious enteric disease spread through food and water contaminated with Salmonella enterica serovar Typhi (S. Typhi). In 2017, more than 10 million cases and more than 110,000 deaths were estimated globally, mostly among children and adolescents in Asia and sub-Saharan Africa. Antibiotic treatment is compromised by the emergence of S. Typhi with resistance to multiple antimicrobials in low-resource settings where suitable diagnosis and treatment may not be readily accessible.
Whole-genome sequencing (WGS), a tool routinely used for infectious disease epidemiology in several high-income countries, is particularly useful to track the emergence and global dissemination of drug-resistant S. Typhi, a human-adapted pathogen that exhibits less genetic variability than other Salmonella serovars. WGS is increasingly being adopted in local and national public health laboratories, but bioinformatics capacity is often a limiting factor in the implementation of genomic epidemiology. Our paper describes Typhi Pathogenwatch, a free application that circumvents this limitation by providing access to genome analysis and actionable information via a web browser.
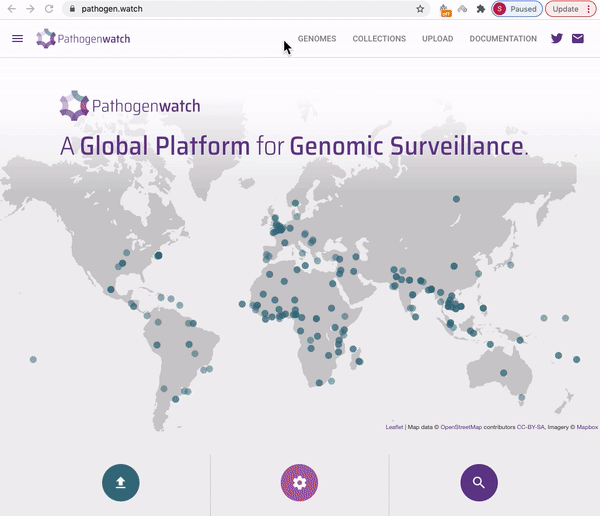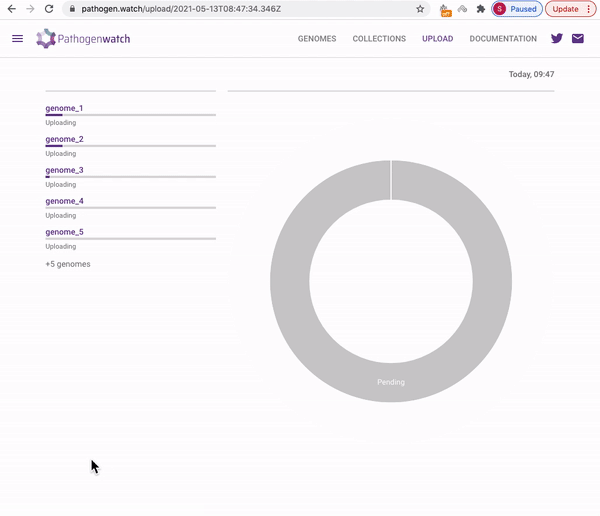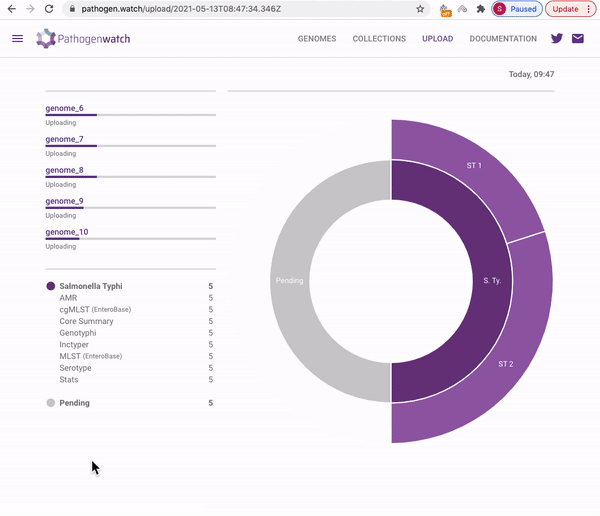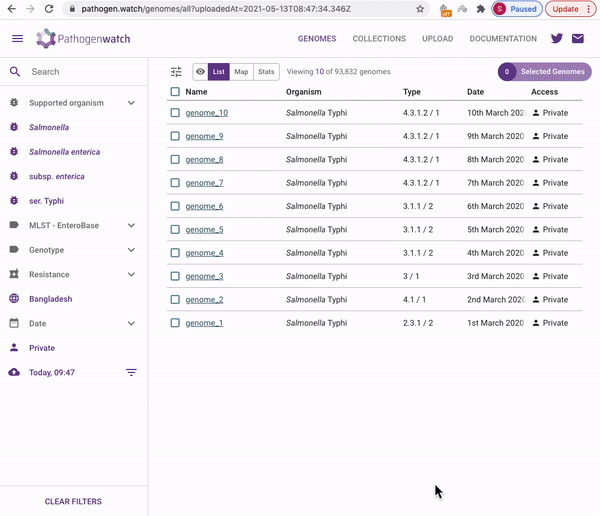
Users interested in exploring public genomes without creating their own collections can browse and gain insights from the public data. The interactive interface links resistance predictions to the tree, map and timeline.
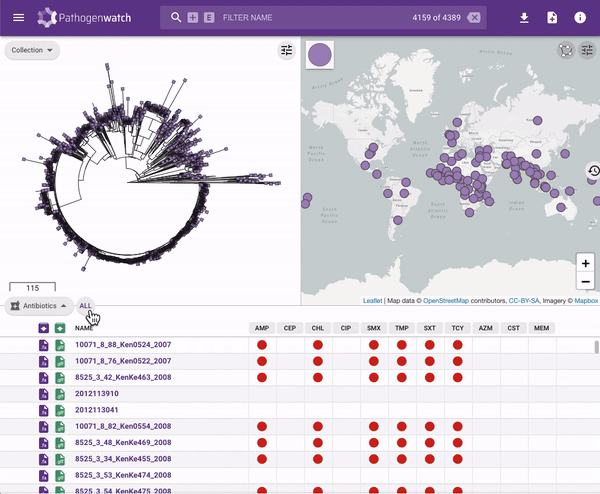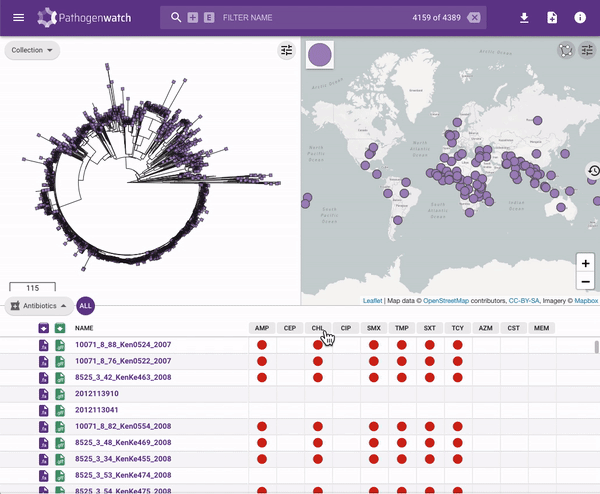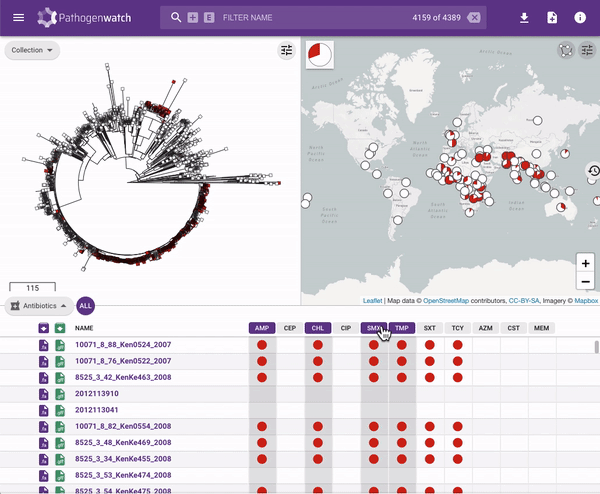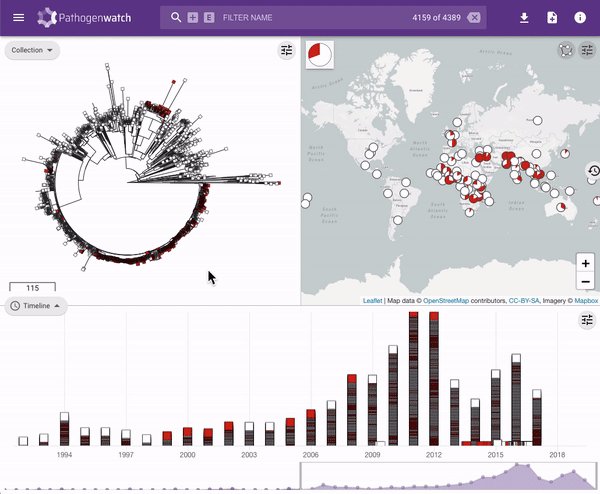
Typhi Pathogenwatch is developed and maintained by the Centre for Genomic Pathogen Surveillance (CGPS) and also incorporates expertise and tools from the typhoid genomics community. We exemplified the importance of this growing resource by showing that when we updated the AMR database with an azithromycin resistance mutation first described in 2019 in Bangladesh and retrospectively searched the >4000 public genomes available in the application we detected the presence of this mutation in genomes isolated in other countries as early as 2007.
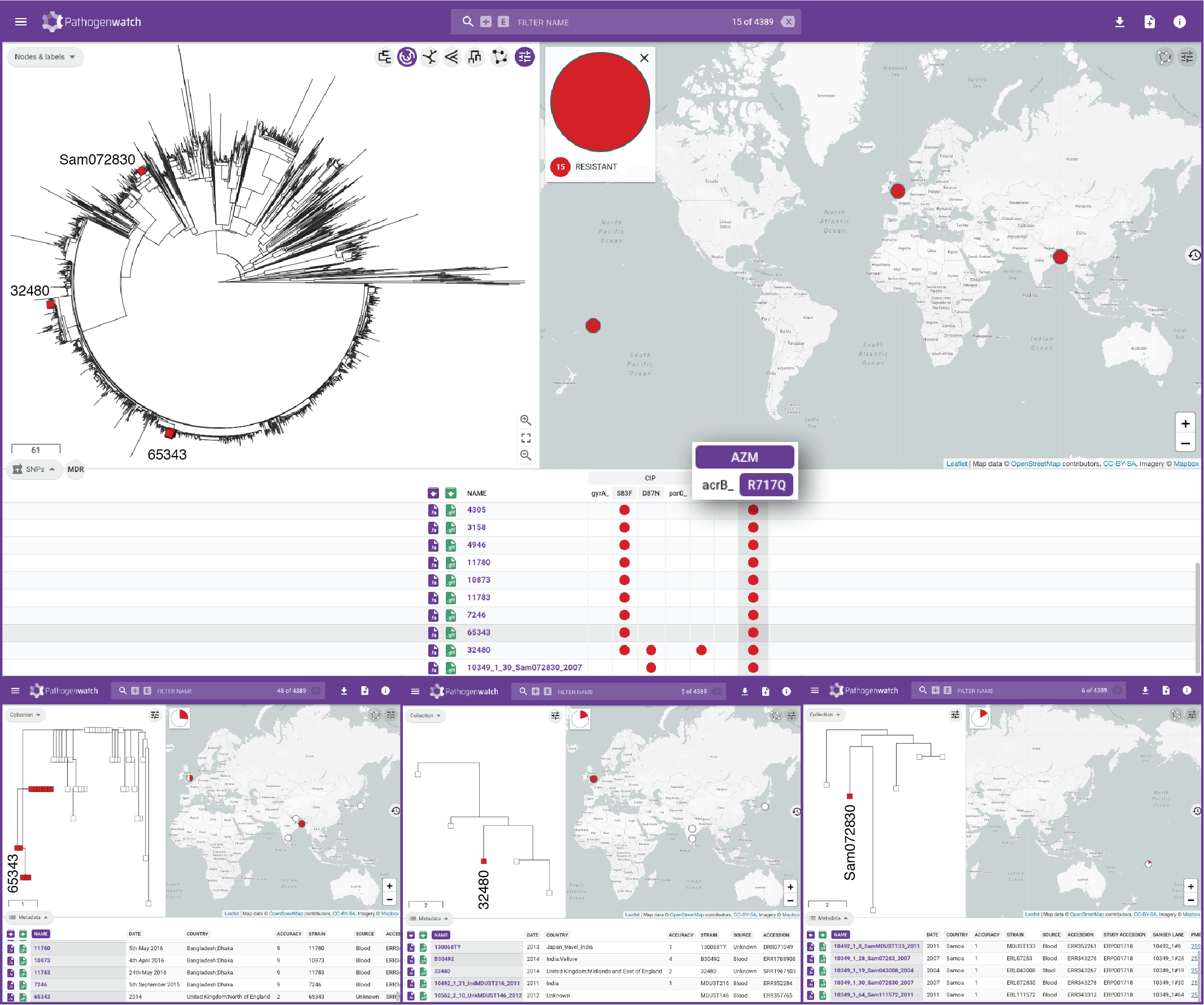
Behind the paper –and the application, there was a substantial effort in curating genome data and linked metadata available both from nucleotide sequence archives and scientific publications. Some of the major obstacles were the lack of raw sequence data deposited for some published studies (i.e., only assemblies available), the lack of harmonization in the metadata reported across studies, and the time elapsed between sequence generation and publication. This, in turn, has recently led us to take the first steps to bring the typhoid genomics community together around more efficient data sharing and management practices to facilitate monitoring the emergence and spread of drug resistance and inform public health action.
The increasing incidence of drug-resistance typhoid has driven efforts to develop new typhoid conjugate vaccines (TCVs). Routine surveillance coupled with WGS can inform decisions on suitable settings for the introduction of vaccination programs and on the evolution of pathogens in response to them. Pathogenwatch linked to routine genomic surveillance around typhoid vaccination initiatives can be used to monitor post-vaccination impact. The consistent provision of patient demographic data would be of particular utility in this context.
Genomics has played a key role in our understanding of the S. Typhi population and dissemination of drug resistance. We believe that the combination of easy-to-use analytics and delivery of public genomes in Typhi Pathogenwatch empowers the public health community and increases the value of the data.
To find out more about Typhi Pathogenwatch you can read the paper in Nature Communications and watch the demo video.